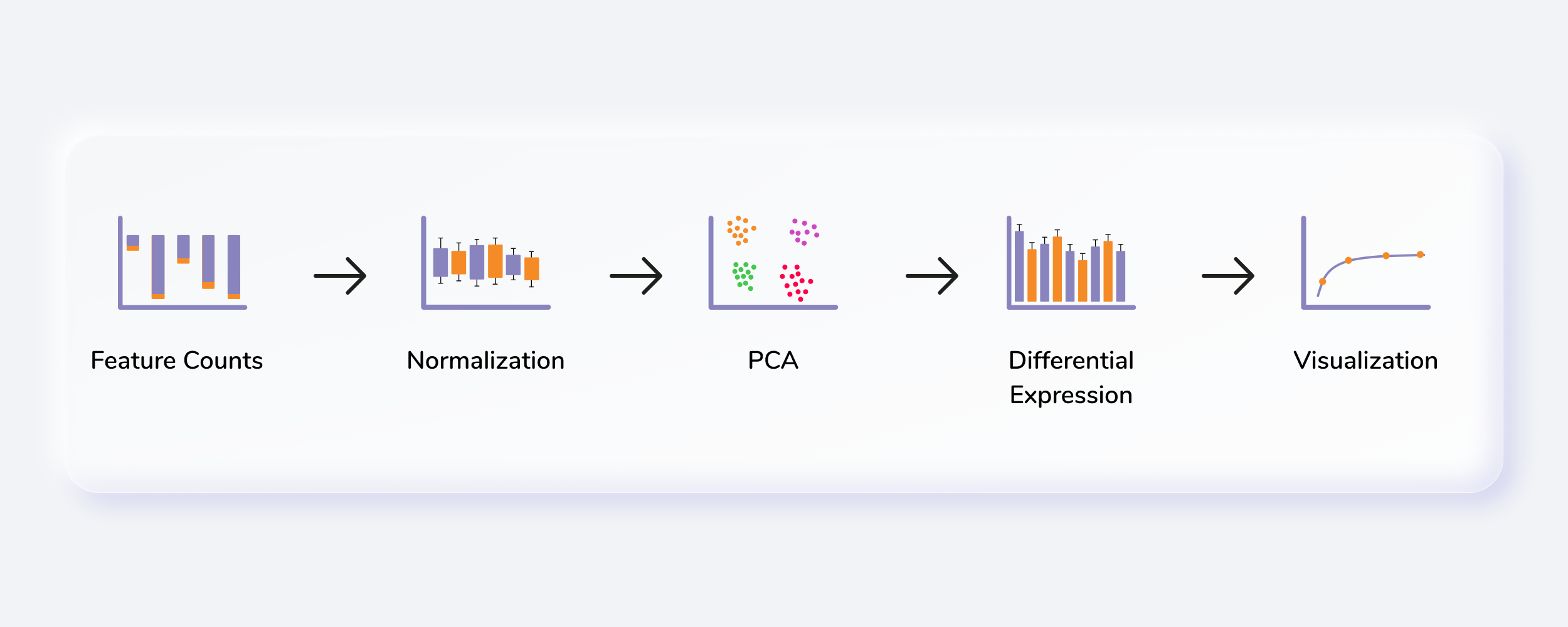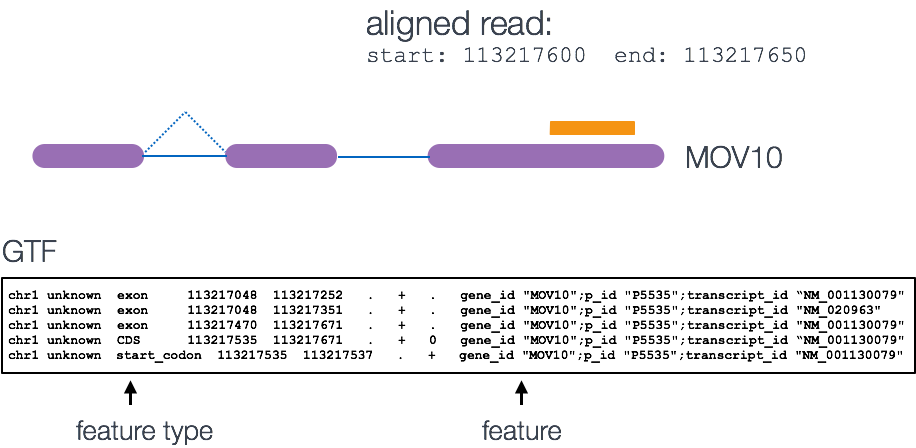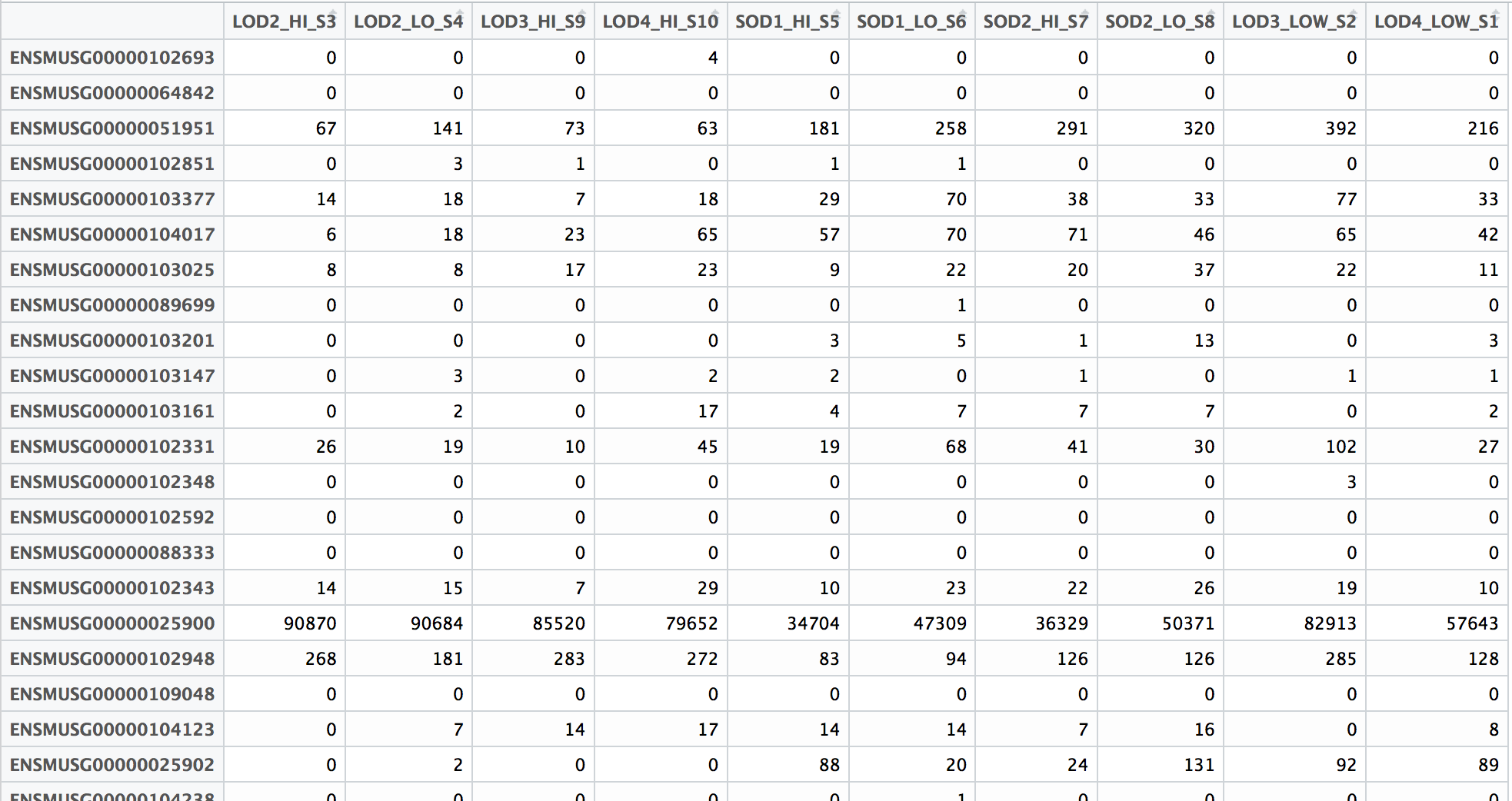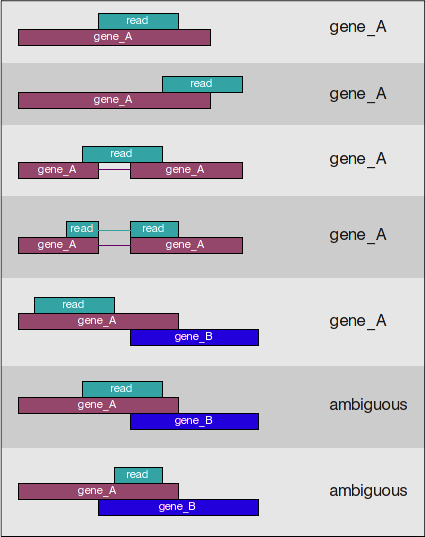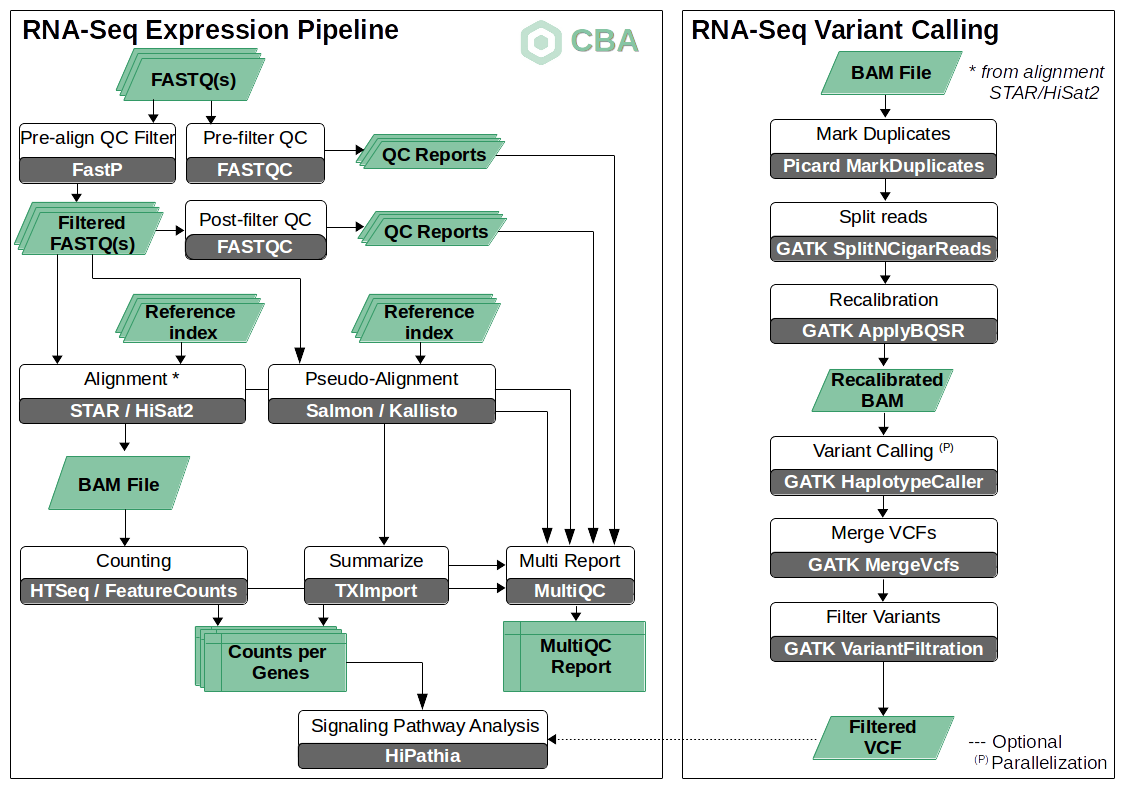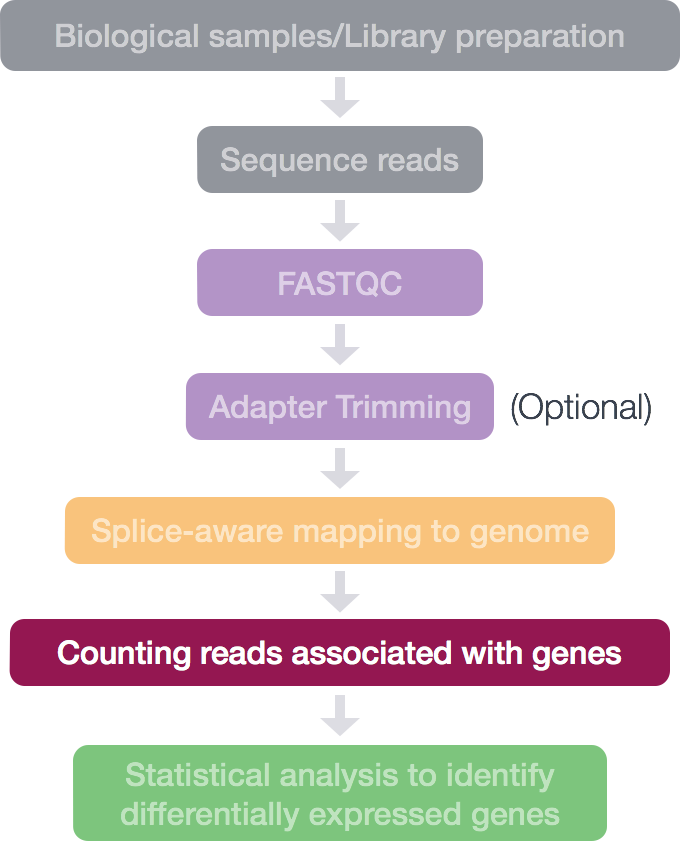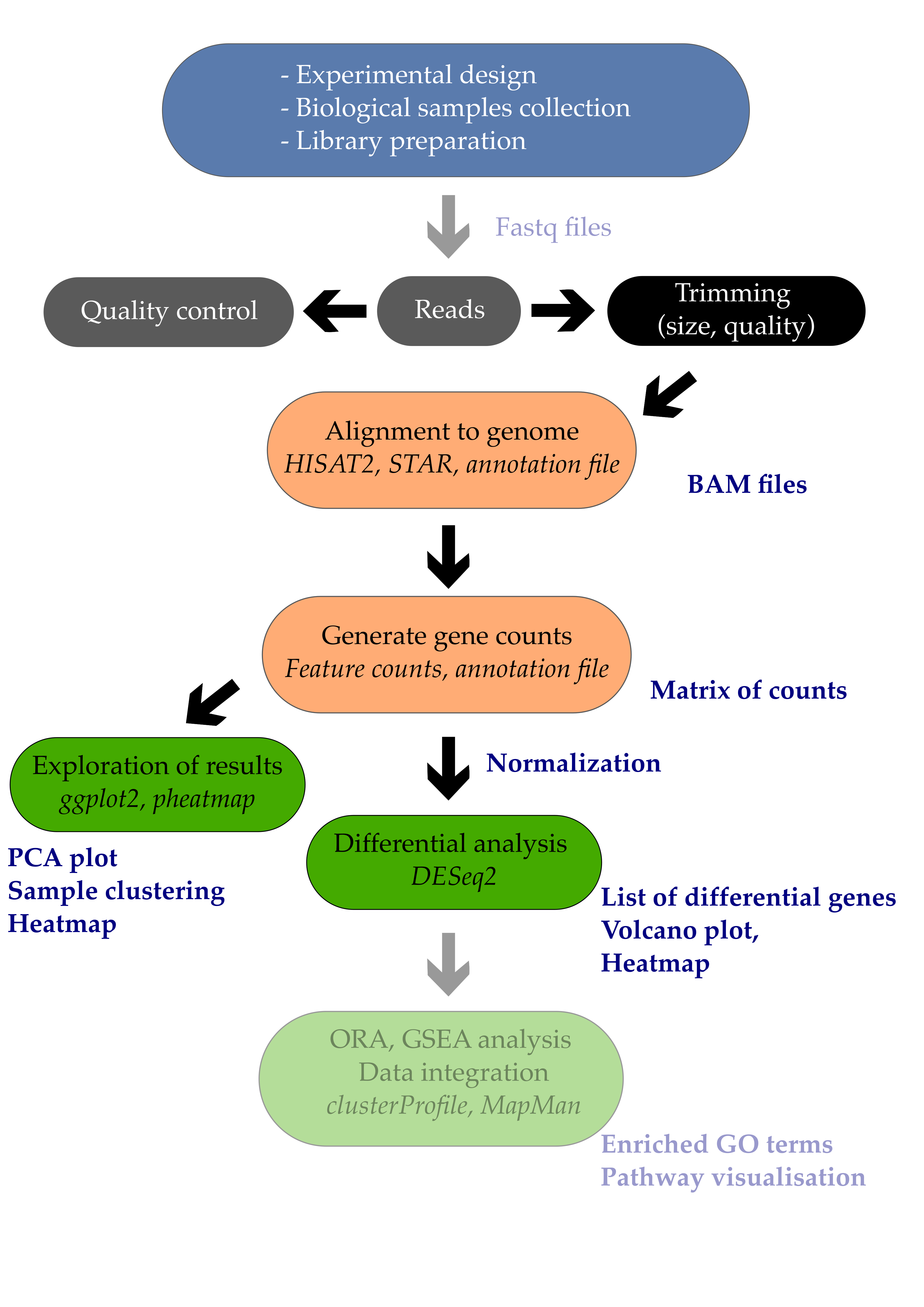
A resource of ribosomal RNA-depleted RNA-Seq data from different normal adult and fetal human tissues | Scientific Data
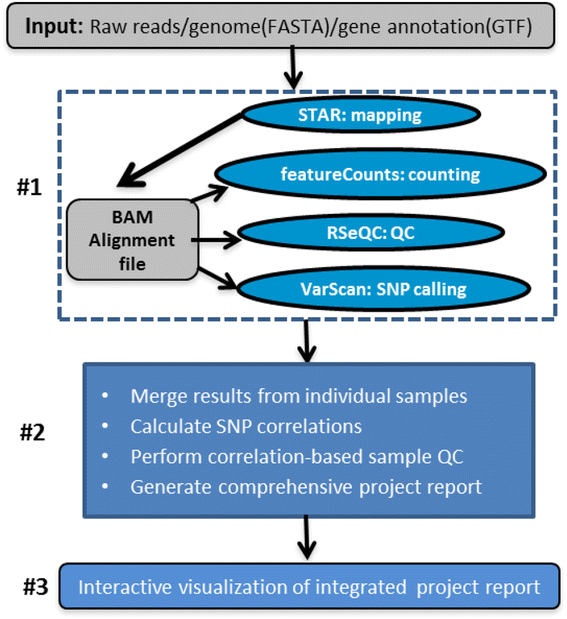
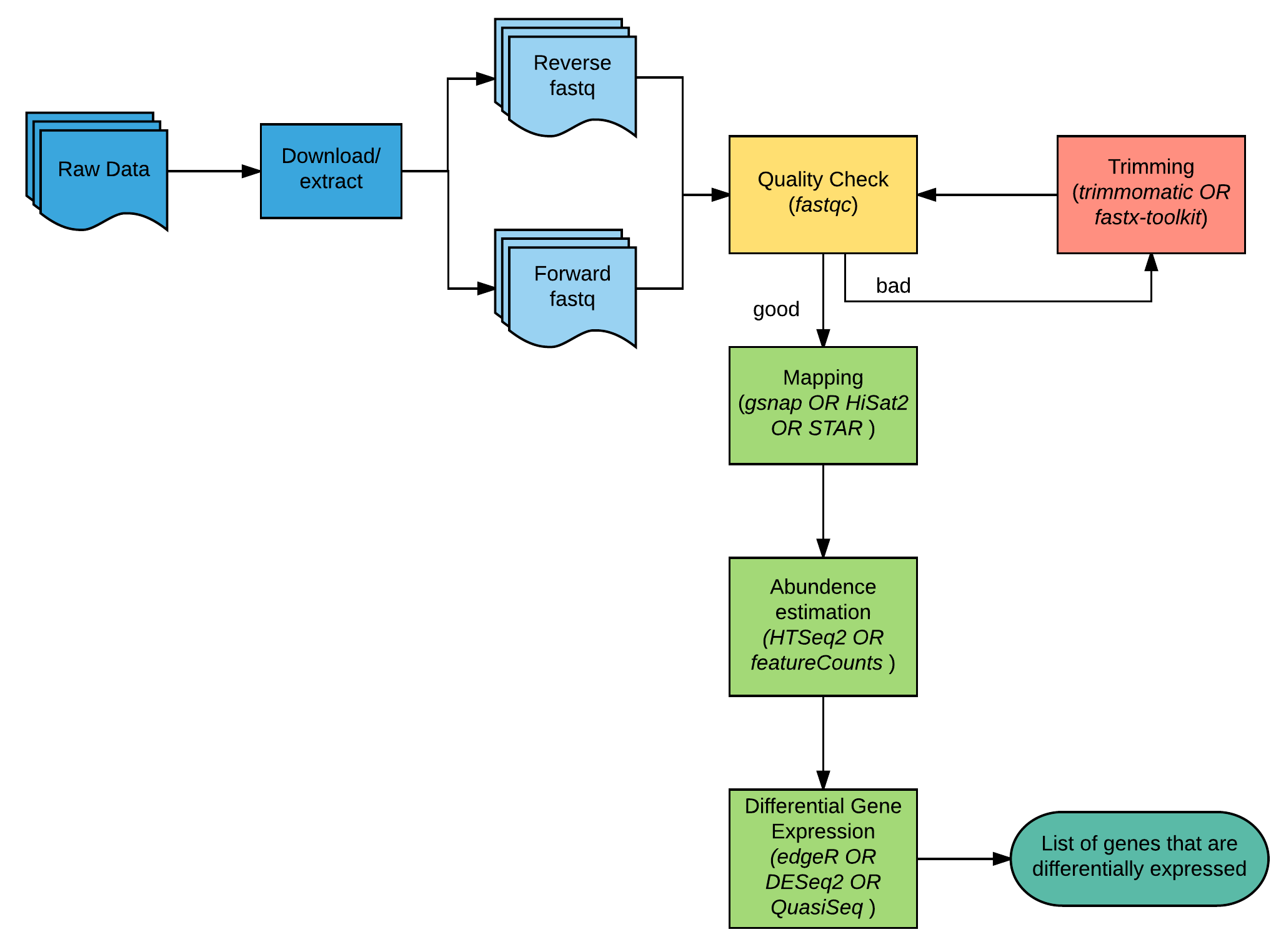
QuickRNASeq lifts large-scale RNA-seq data analyses to the next level of automation and interactive visualization | BMC Genomics | Full Text